Research Projects
Overview
By studying calcineurin, we aim to discover and elucidate new Ca2+-regulated signaling pathways. The calcineurin phosphatase is activated by Ca2+ and calmodulin, and thus dephosphorylates proteins only when Ca2+ signaling is triggered by a hormone, growth factor, neurotransmitter etc. In yeast, we discovered how calcineurin allows cells to survive environmental stress (Goldman et al, 2014). Currently, we are studying human calcineurin which is ubiquitously expressed and plays critical roles in the nervous, cardiac and immune systems (Wigington et al. 2020). We are also mapping signaling pathways and mechanisms for the membrane associated splice isoform, CNAβ1 (Ulengin-Talkish et al. 2021). Calcineurin is best known for activating the adaptive immune response by dephosphorylating the NFAT transcription factors, and is the target of widely prescribed immunosuppressant drugs, FK506 (tacrolimus) and Cyclosporin A. However, these drugs cause many adverse effects due to inhibition of calcineurin in non-immune tissues, where the majority of calcineurin substrates and functions remain to be discovered. Our current strategies to elucidate human calcineurin signaling are described below.
Discovering the human calcineurinome
Protein phosphatases are essential for cell signaling, but have proved difficult to study at the systems-level. We are leveraging recent insights into how calcineurin recognizes substrates to systematically map its signaling network, the calcineurinome, in humans (Wigington et al 2020). Calcineurin interacts with its substrates and regulators by binding to two Short Linear Motifs (SLiMs) termed PxIxIT and LxVP, which occur in intrinsically disordered domains, have low affinity for calcineurin and are degenerate in sequence. These peptides bind to conserved surfaces on calcineurin that are targeted by inhibitors, including the immunosuppressants, FK506 and Cyclosporin A, which block LxVP binding. We leveraged experimental and computational analyses to identify 486 proteins that make up the human calcineurin signaling network or calcineurinome ( see Calcineurin docking motif repository). The network, which is statistically enriched for known calcineurin-regulated processes, also suggests previously undiscovered functions for calcineurin at centrosomes and cilia (Tsekitsidou et al), in neuronal and cardiac signaling as well as nucleocytoplasmic transport, a process whose regulation by Ca2+ signaling was unknown. Discovery of new calcineurin-regulated pathways may help explain the many adverse effects that result from prolonged use of calcineurin inhibitors as immunosuppressants, including onset of diabetes, hypertension, seizures and pain, and provide insights into genetic disorders, such as early onset epilepsy, that are caused by perturbations in calcineurin signaling. We are also working with parents of children that harbor mutations in the calcineurin catalytic subunit, PPP3CA, to identify genetic variants in this gene and to better understand effects of these mutations with the goal of working toward therapeutic interventions.
A pool of calcineurin that localizes to the centriole is revealed by expansion microscopy. See Tsekitsidou et al 2023
Determining the role of calcineurin in regulating nuclear transport
In Wigington et al (2020), we showed that calcineurin co-localizes with components of the nuclear pore basket structure, regulates the phosphorylation of nuclear pore complex (NPC) proteins in vivo and in vitro, and promotes the accumulation of an NLS-containing cargo in the nucleus. Regulation of nuclear transport by Ca2+ and calcineurin is evolutionarily conserved, as yeast nucleoporins, Nup1, Nup2 and Nup60 also contain PxIxIT motifs and are dephosphorylated by calcineurin. Current studies in the lab are using light-regulated reporters for nuclear import and export to identify how calcineurin regulates nuclear transport.
Palmitoylation targets the calcineurin phosphatase to the phosphatidylinositol 4-kinase complex at the plasma membrane
Alternative 3’ end processing of the CNAβ gene (PPP3CB) generates CNAβ1 and CNAβ2: catalytic subunits that differ only at their C-termini and complex with the CNB regulatory subunit to form the calcineurin enzymes, CNβ1 and CNβ2. The CNβ1 C-tail gives this enzyme unique regulation and localization to membranes, where it accesses distinct substrates from other (cytosolic) calcineurin isoforms. We are discovering signaling by CNβ1, which is conserved in vertebrates and widely expressed, but has been little studied.
In vitro, we showed that CNAβ1 has surprising enzymatic properties due to the presence of an autoinhibitory ‘LxVP’ SLiM sequence in its C-terminus that blocks substrate binding and limits Ca2+/calmodulin-dependent activation of the enzyme (Bond et al. (2017)). Our recent in vivo studies (Ulengin-Talkish et al. 2021) show that CNAβ1 is predominantly membrane-associated, in contrast to canonical, cytosolic calcineurin isoforms. This is due to lipidation (palmitoylation) of CNβ1’s unique C-tail which is dynamic due to depalmitoylation by ABHD17A. We also identified proteins that interact preferentially with CNAβ1 compared to the related CNAβ2 (canonical) isoform, which are predominantly membrane proteins, including a highly conserved protein complex (PI4KIIIa, TTC7B, FAM126A/Hyccin, and EFR3B) which synthesizes phosphatidylinositol-4-phosphate (PI4P), a precursor of the critical signaling phospholipid, PI(4,5)P2, at the plasma membrane. Palmitoylation is required for specific interaction of CNAβ1 with this complex. Thus, dynamic palmitoylation confers unique localization, substrate specificity and regulation to CNAβ1. During sustained signaling through G-protein coupled receptors that activate Ca2+ signaling, the PI4P-kinase complex is stimulated to replenish PI(4,5)P2, which is required for sustained signaling. We discovered that calcineurin inhibitors decrease PI4P production under these conditions, identifying a new calcineurin regulated signaling pathway that is likely regulated by CNAβ1. Our studies also suggest there are additional substrates for CNAβ1 in humans and that CNAβ1 activity may be regulated primarily through a unique palmitoylation/depalmitoylation mechanism. We are continuing these studies and identifying the palmitoyltransferases that regulate this isoform.
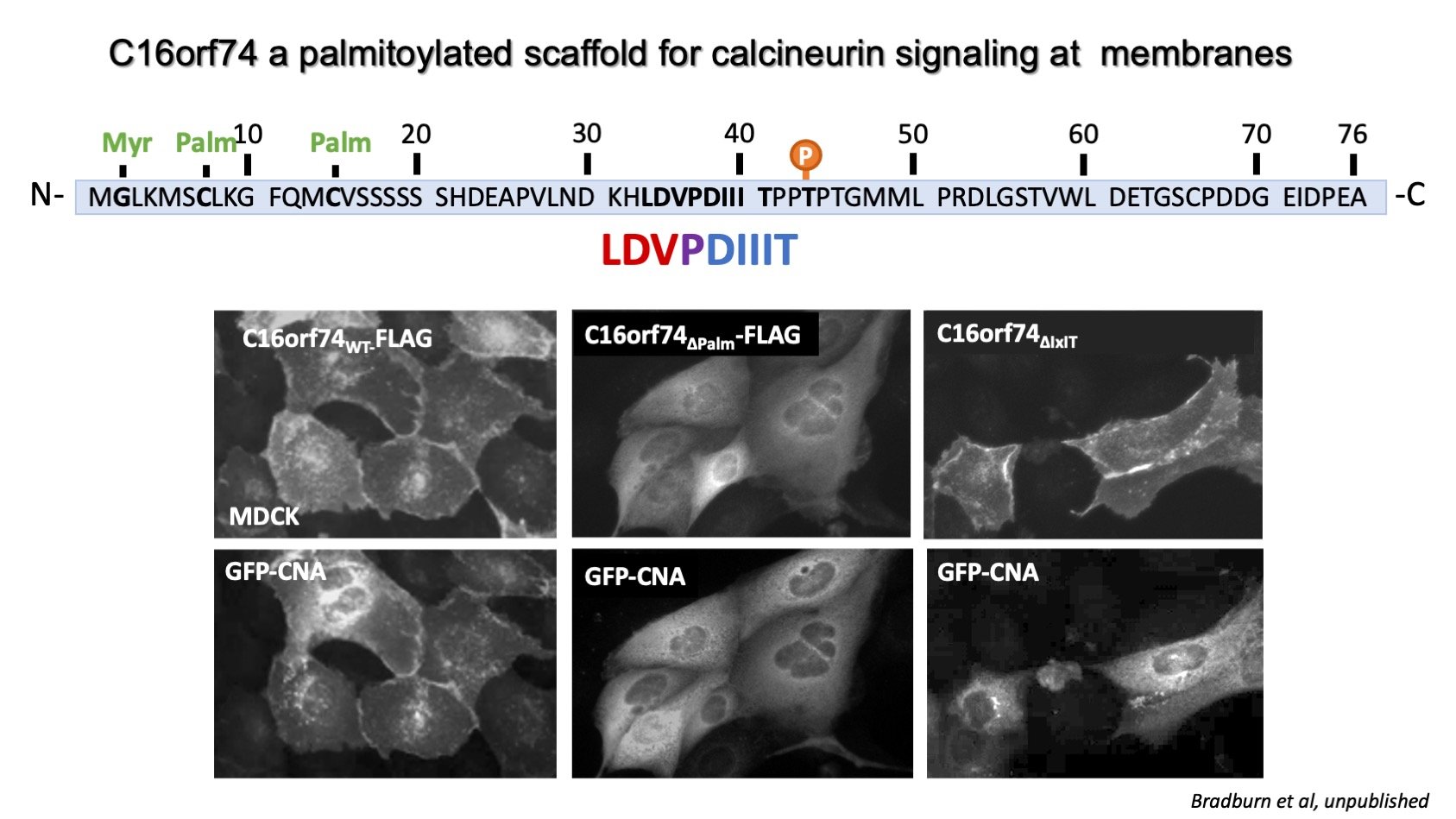
Calcimembrin/C16orf74 targets calcineurin to signaling microdomains at membranes
For calcineurin signaling to occur, it must be co-localized with its substrate(s) to a calcium microdomain, which generally occur in proximity to a calcium channel or transporter at a cellular membrane. See our recent review, A cellular atlas of cacineurin signaling for more information about calcineurin’s subcellular distribution (Ulengin-Talkish and Cyert, 2023). We are studying a small, disordered, uncharacterized protein, C16orf74, which targets calcineurin to membranes utilizing an unusual composite LxVPxIxIT SLiM. See Wigington et al for other instances of this composite motif. C16orf74, which we named calcimembrin, associates with membranes via lipidation, especially reversible palmitoylation. It is also dephosphorylated by calcineurin using a unique mechanism that requires two C16orf74 molecules per calcineurin allowing one C16orf74 to bind calcineurin via its PxIxIT and the other C16orf74 to undergo LxVP-dependent dephosphorylation. High levels of C16orf74 correlate with poor prognosis for melanoma, pancreatic and head and neck cancer. We are currently investigating how C16or74 directs calcineurin signaling using structural, biochemical and cell biological assays. See our paper about calcimembrin which is in press at Nature Communications and was a collaboration with the Arthanari lab at Dana Farber Cancer Institute.
Mapping calcineurin signaling pathways that mediate pancreatitis
Pancreatitis is an excruciatingly painful medical emergency caused by inflammation of the pancreas, a banana-shaped organ that is located behind the stomach and that produces insulin and a slew of pancreatic digestive enzymes. There are no targeted therapies for pancreatitis, and its causes are not well understood. However, the disease state is caused by a prolonged pathological rise in Ca2+ that induces calcineurin signaling, and calcineurin has been shown to be critical for the development of pancreatitis in work from the lab of Sohail Husain, Department of Pediatrics/Gastroenterology, Stanford Medicine. We are currently collaborating with the Husain lab to systematically identify signaling pathways and phosphoproteins that are regulated by calcineurin during pancreatitis using unbiased phosphoproteomic analyses. By identifying calcineurin substrates and signaling pathways in pancreatitis we aim to uncover fundamental information that will aid in developing focused pancreatitis therapies in the future.